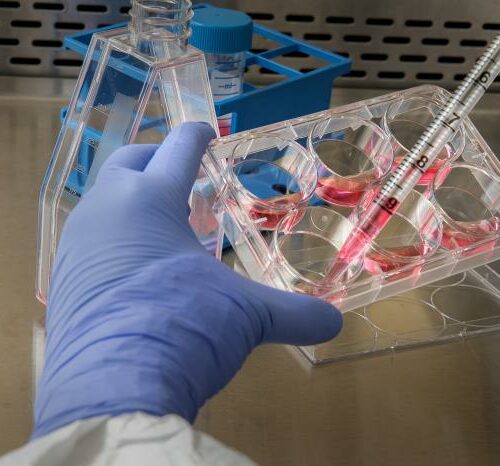
Some critters in the ocean are reclusive, hiding from human probes and trawls. Other critters are rare, driven close to extinction from warming and increasingly acidic waters.
Studying rare and reclusive creatures has posed problems for scientists in the past. In recent years, environmental DNA, or eDNA, has helped. To isolate eDNA, scientists scoop water from the ocean.
“You’re capturing stuff that is falling off the fish: scales, slime or poop, and you’re getting tiny little tiny fragments, individual cells and whatnot. Then, you’re trying to find those cells in a giant bottle of water,” said Andrew Shelton, a research ecologist with the National Oceanic and Atmospheric Administration.

Aboard the Bell M. Shimada, Jennifer Fisher, left, and Rebecca Smoak collect water samples that were collected at different ocean depths.
On a two-week long voyage this May, scientists took samples while on board NOAA’s Bell M. Shimada research vessel during a survey of the Northern California Current ecosystem.
“One person will collect eDNA water samples and come to this station,” said Jennifer Fisher, co-chief scientist on the Shimada, as she showed others how to use the eDNA filtering equipment.
After sampling various ocean depths, the scientists ran water through silver-dollar sized filters, which looked like tiny coffee filters.
The hum of the pumps clouded other noise in the ship’s wet lab, where scientists spent much of their nearly two-weeks aboard the ship, collecting data that could help shed more light on the tiny plankton and other ocean life that serve as food for salmon.
The filtration process was sometimes painstaking and time-consuming, depending on the amount of gunk in the water. Pumps pulled the water through glass cups covered with aluminum foil. Then, the excess water flowed through pipes and into a larger bottle.

Talia Davis filters water to collect environmental DNA, or eDNA, aboard NOAA’s Bell M. Shimada.
Scientists carefully removed the filters, which they hoped were full of eDNA. The scientists used tweezers to put the filters in plastic tubes to be analyzed in a land-based lab.
Over the past decade, eDNA technology also has allowed conservation scientists to figure out whether rare and endangered species are present in particular environments. More recently, fisheries managers are using eDNA as a tool to learn more about fish stocks in vast spaces, such as the ocean.
Around 65 of eDNA samples scientists collected on the Shimada went to Nick Adams, a research oceanographer with NOAA in Seattle. Survey expeditions later in the season also collected samples for Adams to analyze.

Scientists used tweezers to fold filters full of eDNA samples into quarters. They then placed the filters into test tubes.
Adams hoped to determine the diversity of phytoplankton in the water samples collected on the Shimada. Phytoplankton are the first link in the oceanic food web. Phytoplankton feed other organisms that salmon eat.
“When we do our fancy analysis, we’ll be able to say, ‘Oh, this piece of DNA belongs to this organism, this one belongs to this organism,’” Adams said.
Then he’ll work with other scientists and data collected from the Shimada’s survey to compare information and get a bigger picture of what was going on in the ocean where the samples were collected.
About three months after the Shimada’s May 6-17 trip, Adams awaited the analysis of the eDNA samples. Scientists had collected samples for Adams at each stop the vessel made.
Adams stored those samples in a freezer at -80 degrees Celsius, or -112 degrees Fahrenheit.

Anna Bolm collects eDNA samples aboard NOAA’s Bell M. Shimada. On the May 6-17 trip, scientists collected around 65 eDNA samples for research oceanographer Nick Adams.
To prepare the samples, he stuck the filters in a tube with tiny glass beads and a DNA extraction buffer. Then a bead beater machine shook the samples.
“It just beats the snot out of them,” Adams said. “Just to get all the little particles, and especially plankton, that I’m looking for. Some phytoplankton have these silica shells, and they’re hard to break open.”
That’s why the bead beating machine comes in handy. Adams then stored the samples overnight, incubating them at around 132 degrees Fahrenheit.
The next morning, Adams spun the sample tubes and removed the DNA extraction buffer.
“DNA will stick to glass under high salt concentrations. So the DNA will stick to the glass tubes. You wash away all the bad stuff and then you use water or another kind of buffer to strip the DNA off the glass. And then you have purified DNA,” Adams said.
The purified DNA is then examined in a process called DNA sequencing.
A high-powered computer can compare the DNA sequences to databases of known phytoplankton DNA sequences, which will hopefully give Adams information on the diversity of plankton DNA in the samples collected on the Shimada. The scientists hope that information could indicate if there will be good food available for salmon in the ocean.
Shelton said there is a lot of promise in what eDNA could do for science in terms of conservation and fisheries management. However, there is still a lot of room for eDNA to grow, he said.
“It’s very hip,” Shelton said. “It’s sort of a new kid on the block right now. Whenever there’s a new kid on the block, the new kid gets a lot of attention and a lot of money.”
The science of eDNA is still in its infancy, but it will become even more relevant in the years to come, Shelton said, helping to complement other types of fish surveys, such as trawling.
In fisheries, these other types of surveys have gone on for decades. Now, as scientists collect and safely store eDNA data, that information will continue to give scientists an idea about the concentrations of fish in the ocean, Shelton said.
“In five years the data will be very useful. In 10 years it will be very useful. In 20 years it will be very, very useful,” Shelton said.
Recently, Shelton wanted to see if eDNA could enhance fisheries management on a large scale. Previously, eDNA mostly had been put to use by conservationists.
Shelton used eDNA to survey the Northern California Current for hake, the largest commercial fishery off the West Coast. The data from the eDNA showed similar results to more traditional acoustic and trawling surveys.
But, the ocean is vast. Surveyors could be feet away from eDNA samples, but the currents could push the eDNA in the opposite direction. One way to prevent that, Shelton said, is to take a ton of samples.
Scientists can’t infer a universe of data from one sample, Shelton said.
Think about it similarly to looking for fish in Seattle’s Green Lake, he said.
“You don’t go out and stick your dip net in and be like, ‘Zero fish in my dip net. There must be zero fish in Green Lake,’” Shelton said. “You would sample all around the lake, in the shallow water and in the deep water. You’d do a systematic, thoughtful search.”
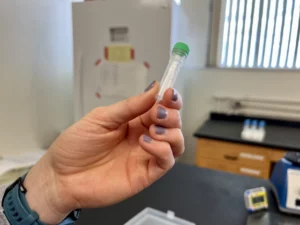
Meghan Parlsey shows off eDNA samples she’s collected for her doctoral work at Washington State University.
Across the state in Pullman, Meghan Parsley has collected eDNA samples for her doctoral work, one part of which involves using the quantity of eDNA to estimate the population size of wood frog tadpoles in Connecticut.
“This is where the magic happens,” Parsley said, walking into a sparse, clean lab at Washington State University.
Keeping unwanted DNA out of the lab is tough and involves a lot of bleach. “I have lots of bleach stained clothes,” Parsley said.
All that bleach helps ensure stray DNA from pretty much anywhere doesn’t mix with the tadpole eDNA she wants to process.
“I like to relate it to baking, right? There’s a recipe and instructions. You have to follow it pretty closely, but it’s really just a series of chemical reactions where you’re kind of breaking apart the cells to have access to the DNA,” Parsley said.
The Goldberg Lab at Washington State University, where Parsley is a lab member, often focuses on rare species with low populations.
“I think eDNA is really changing how people think about monitoring and finding where things are,” she said.
In fact, she’s working to push the envelope further, studying eRNA. If DNA is the blueprint of a body, she said, RNA translates that blueprint into action.
While Parsley said eRNA is more finicky than eDNA, she said she wants to use eRNA to help determine the age of different bullfrogs and tiger salamanders. One drawback of eDNA right now is that it can help measure the amount of something instead of helping scientists learn more about age, Parsley said, which she believes eRNA will be able to measure.
Another potential future area of research scientists hope to study could use eRNA to measure the stress level of a particular species, Parsley said.
To determine age with eRNA, she’s trying to detect tadpoles among a population of adult bullfrogs, which are invasive in the West.

Meghan Parsley is studying eDNA and eRNA for her doctoral work at Washington State University.
This type of information could help land managers who might spend days or weeks trying to find a bullfrog that they heard in the distance. Sometimes those frogs are just passing through, Parsley said, sometimes the frogs are reproducing in a nearby pond.
If land managers submitted a sample of water that scientists could use to see if bullfrog tadpoles were present, it could save a lot of time and effort, Parsley said.
“We’re trying to help discern if a frog is reproducing without having to spend a bunch of time stalking this one frog that someone heard,” Parsley said.
While eDNA or eRNA won’t fix every conundrum in conservation science, it could provide another tool to help land managers in the future, she said.
“Why not use eDNA and save our manpower and lots of hours and be able to spread the love to other projects and other species,” Parsley said.
In addition, Shelton said eDNA could make science more equitable. Scientists don’t need a fleet of vessels to stay out on the ocean for days or weeks. The technology could open up studies for wildlife in parts of the world that don’t have as much access to research dollars, Shelton said.
“There’s this opportunity for spreading a lot of this information and using it in places that are not as well developed,” Shelton said.